@_rossry Hi, thanks for considering funding the project! In response to both of your comments, here's a more detailed look at the cost budgeting.
TLDR: the $15k projection is our highest expected cost and was chosen to maximize likelihood of success. In reality, cost may be lower once budget is optimized, but hard to predict ahead of time without expert guidance, and securing funding would make it much easier to get that guidance. If costs are lower than raised amount, project can be easily expanded to have more impact. If raised amount is <15k/amount needed to benchmark 30 genes and push industry adoption with GenScript, there are meaningful goals we can achieve by benchmarking 3-10 genes like publishing in a top tier journal to push research community adoption.
Detailed response:
Here is the benchmark sequence database we had planned to use for testing: https://github.com/Lattice-Automation/icor-codon-optimization/tree/v1.4/benchmark_sequences/dna These were selected based on application and use in other research papers which would allow for easier comparison between the ICOR approach and others, and so we don't need to reinvent the methodology. Average BP ~1800. We looked at production via GenScript. One important factor to note is that we need to have the full process done externally from Gene Synthesis, not Plasmid Preparation. This includes steps of adding antibiotic resistance (to select out bacteria which successfully took up the plasmid), adding excision sites around the sequence to insert it into the plasmid, etc. Theoretically, we could do this ourselves which would reduce cost. But, this gives the benefit of being robust, standardized, and would save resources in lab materials & support. Taking this into account, we can leave the rest of the settings default. Ideally, I would prefer to add additional checks like ensuring sterility, producing it without animal products, etc but let's leave it at the bare minimum. Also, note that ~1/3 of the sequences are "Complex" so GenScript is unwilling to give a full cost estimate until they are sure we are ordering it.
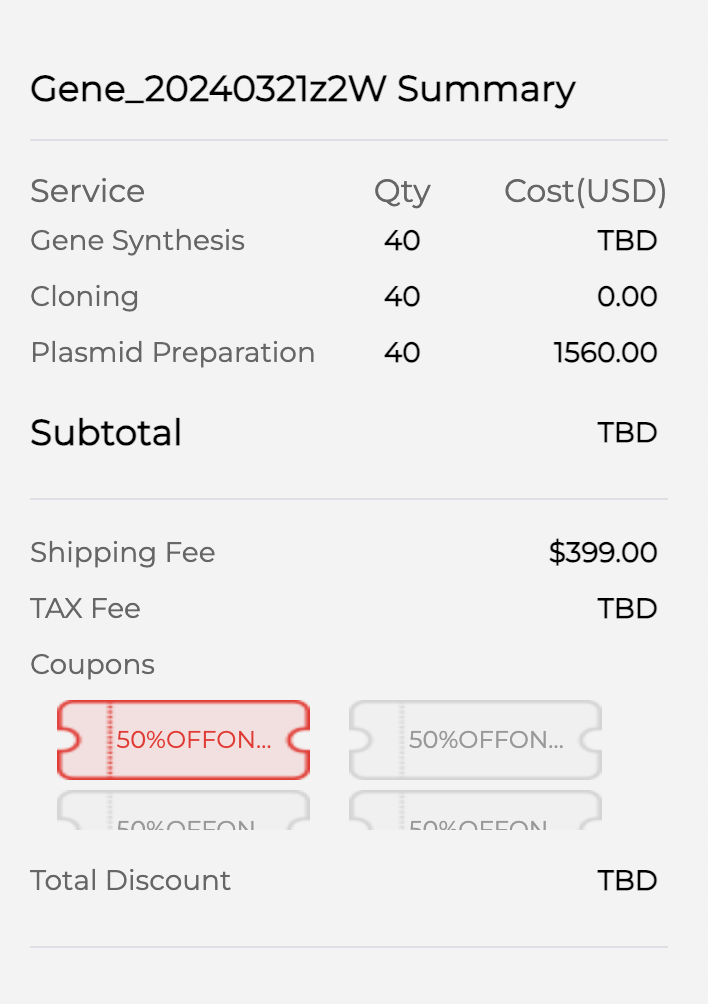
I have not requested a full quote as I need to verify some parts of the methodology. But with 40 sequences, the preliminary cost is given as $1960 (shipping to Boston, MA). This comes to $50 per sequence. Now, this cost would actually be higher as they have not added in gene synthesis cost, but with student discount, the bulk discount as we are actually ordering 2-3x this, and institutional discounts, I ball-parked that the cost would stay at around $50/sequence. I understand this is not the most detailed analysis for cost and there may be other competitors which can give us a cheaper rate. If we are funded, my plan is to speak with some PIs and mentors with more wet lab experience who can help me figure out how to optimize cost. I apologize for not having the most detailed budget, but rest assured that I will work with experts on this and if it can be done for cheaper, would use the remaining funds to expand the outcomes (eg: test in multiple host organisms which is becoming more relevant for some specialized drugs)
As for your second question. There are definitely meaningful goals we can achieve with reduced funding! Most published papers which seek to show a new method for optimizing protein expression use far fewer benchmark genes - usually 3-10.
(For example: https://www.nature.com/articles/s41586-023-06127-z#Sec10, for COVID mRNA production and you can see many others in recent research with a similar amount of test sequences. Also side-note: they are also initially evaluating their tool with CAI which we did in our publication and they look for this "sweet spot" in CAI with a human-made computational algorithm below peak CAI due to the problem of too few tRNAs mentioned in the proposal. This is a cool approach, but I suspected our AI approach outperforms this, and this paper was in Nature (!) in 2023 so it's very state of the art).
So with $5k we could test approximately 10 benchmark genes, $3k would still let us do 6, etc. This would result in a paper publication that I expect would be high impact for the research community, would solve the main criticism of ICOR which was that it was not wet lab tested, and would be a major stepping point towards adoption. However, it would not meet GenScript's metrics for adoption in their pipeline and I think it would increase the timeline for it to reach industry.
Grateful for your consideration!